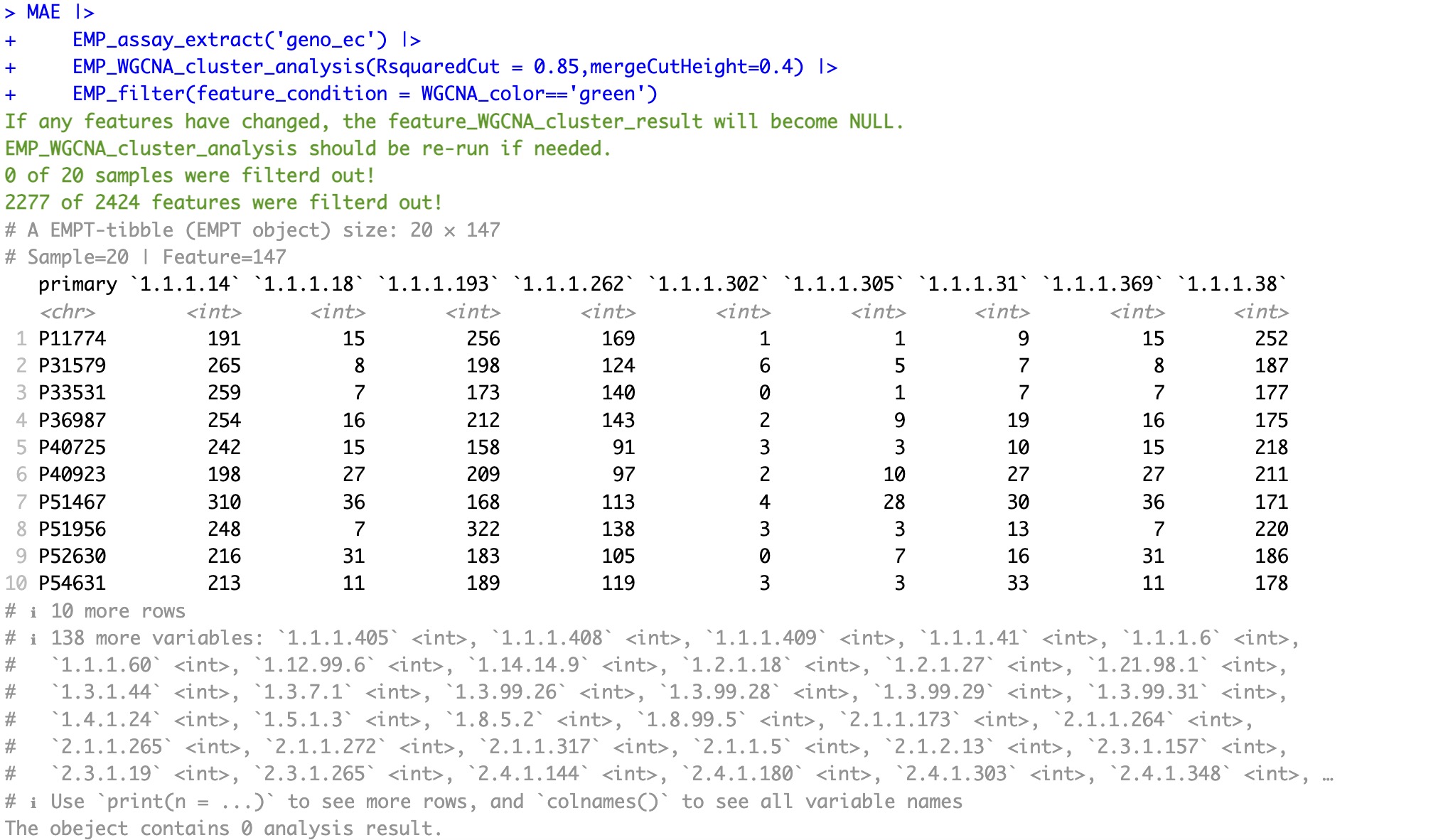
6.9 EMP_WGCNA_cluster_analysis
Weighted correlation network analysis (WGCNA) aims to discover co-expressed gene modules, explore the association between gene networks and phenotypes of interest, and find the core genes in the network.
6.9.1 WGCNA Clustering Module
This module integrates the relevant modules of the WGCNA package, which can quickly complete clustering based on the WCGNA algorithm.
🏷️Example:
MAE |>
EMP_assay_extract('geno_ec') |>
EMP_WGCNA_cluster_analysis(RsquaredCut = 0.85)

Note:
The parameter
The parameter
EMP_WGCNA_cluster_analysis inherits the usage of the function WGCNA_blockwiseModules from WGCNA package, and help adjust the clustering effect.
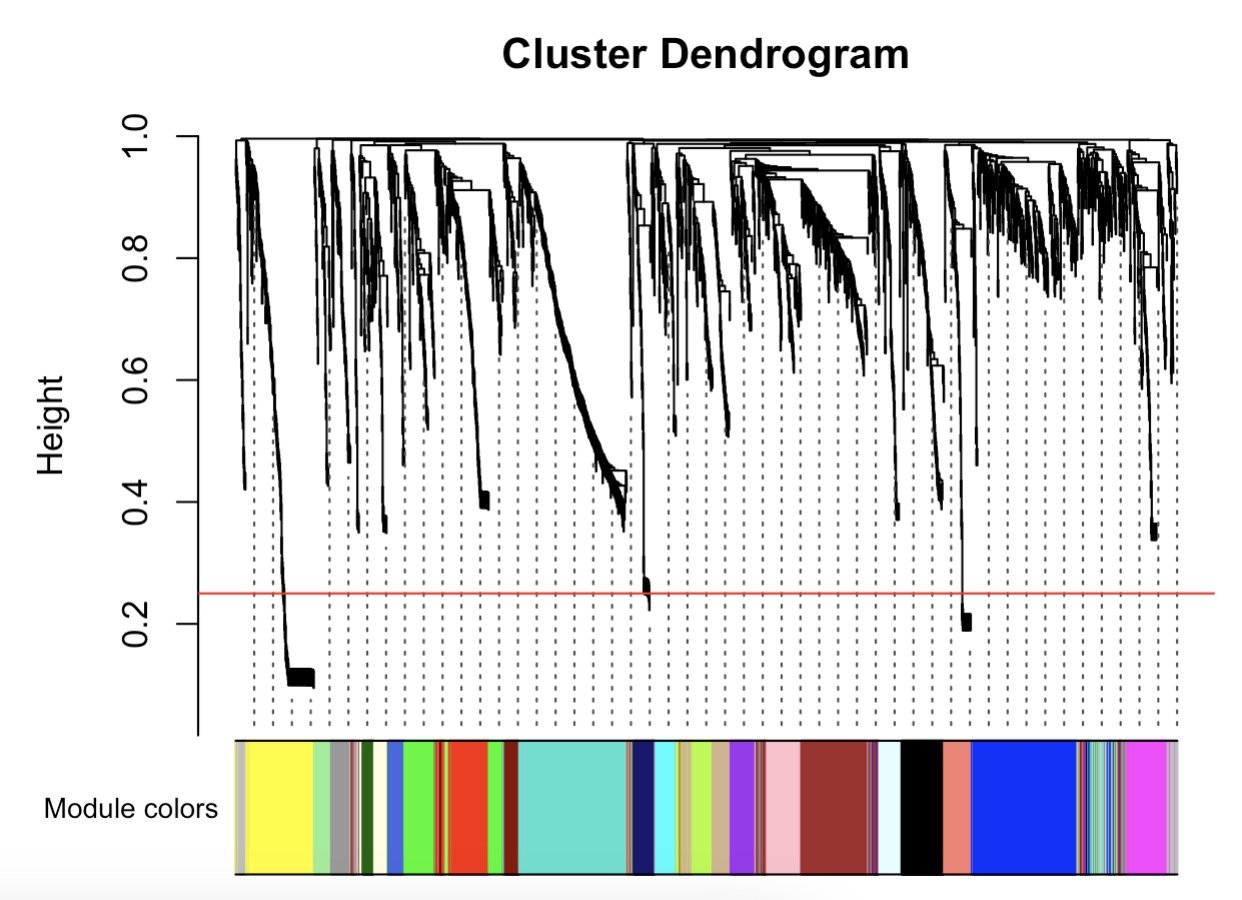
MAE |>
EMP_assay_extract('geno_ec') |>
EMP_WGCNA_cluster_analysis(RsquaredCut = 0.85,mergeCutHeight=0.4)

6.9.2 Filtering the WGCNA clustering result
The WGCNA cluster analysis results can be filtered using the module EMP_filter. This process has been widely used in subsequent enrichment analysis.
🏷️Example:
MAE |>
EMP_assay_extract('geno_ec') |>
EMP_WGCNA_cluster_analysis(RsquaredCut = 0.85,mergeCutHeight=0.4) |>
EMP_filter(feature_condition = WGCNA_color=='green')